
Research

Am not I
A fly like thee?
Or art not thou
A man like me?
- William Blake, The Fly
A comparison of the molecular mechanisms governing heart development in vertebrates and Drosophila reveals a remarkable conservation of all major regulatory components, including both signals and transcription factors. Furthermore, many of these conserved cardiac regulators are found to be mutated in various types of human congenital heart disease. The identification of conserved genes involved in cardiac development in both Drosophila and mammals, and detailed investigations of their functions will thus provide considerable insight into related mechanisms controlling cardiogenesis in vertebrates, including humans.
The Ahmad Lab therefore utilizes integrated genetic, genomic, and computational strategies to identify novel genes involved in both Drosophila and mammalian heart development, and pursues detailed investigation into both their function and regulation. Our ultimate goal is to arrive at a comprehensive understanding of heart development which incorporates both a systems-level view of the underlying genetic network and detailed examinations of the roles of individual genes.
We also use similar integrated approaches to examine and dissect biological processes involved in the development of other organs besides the heart. Current research projects in the Ahmad Lab include:
Functional analysis of cardiogenic processes regulated by Forkhead/Fox transcription factors.
At least eight transcription factors (TFs) of the Forkhead/Fox family are required for proper cardiac development in mammals, while mutations in four Fox genes have been linked to human congenital heart defects. However, relatively little is known about the molecular mechanisms or the downstream targets by which these Fox-mediated developmental functions are brought about. Our prior work in Drosophila identified the roles of two Fox genes, jumu and CHES-1-like, in specifying cardiac cell subtypes and number by mediating cardiac progenitor cell divisions, and in determining cardiac fates. We also showed that Fox TF binding sites were significantly enriched in the enhancers of cardiac genes. Collectively, these results indicate that these two Fox TFs integrate multiple cardiogenic processes by regulating a large number of downstream target genes, raising the question of what these target genes are and what their individual functions might be during heart development.
Utilizing genome-wide transcription expression profiles of mesodermal cells from wild-type embryos and embryos with appropriate genetic perturbations in the Fox genes, we have identified 963 jumu-regulated and 427 CHES-1-like-regulated target genes. We are now identifying and functionally analyzing the diverse cardiogenic cellular processes mediated by these Fox targets.
Determining the functional roles of regulatory genes implicated in cardiogenesis.
By combining expression profiling, statistical meta-analysis and large scale in situ hybridizations, we identified novel Drosophila regulatory genes expressed in the heart or its precursor, the cardiac mesoderm. In a separate project, using machine learning to classify cardiac enhancers based primarily on the transcription factor binding sites of orthologous vertebrate TFs, we identified additional Drosophila TFs regulating cardiac genes. And a third project by our colleagues identified putative mammalian cardiogenic genes from epigenetic gene expression signatures of mouse and human embryonic stem cells differentiating along the cardiac lineage.
Integrating results from these three distinct projects has enabled us to identify many cardiogenic regulators conserved between Drosophila and mammals, with their conservation suggesting potentially central roles in heart development. Utilizing both Drosophila as a model system and mammalian stem cells directed to differentiate along the cardiac lineage, we are now examining the functional roles of these conserved regulatory genes in cardiogenesis in greater detail.
Understanding the regulatory processes giving rise to distinct cardiac cell subtypes.
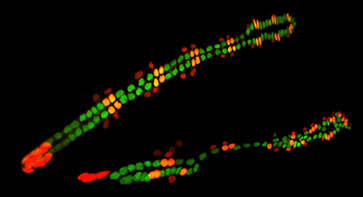
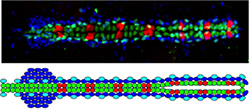 In both Drosophila and mammals, the heart is comprised of many different cell types, raising the question of how these distinct cardiac cell subtypes are specified and differentiated. Taking advantage of the unique expression profiles and stereotypical metameric positioning of the eight different cardiac cell types in Drosophila at single cell resolution levels, we are investigating the signaling and transcriptional processes that determine their fates.
In both Drosophila and mammals, the heart is comprised of many different cell types, raising the question of how these distinct cardiac cell subtypes are specified and differentiated. Taking advantage of the unique expression profiles and stereotypical metameric positioning of the eight different cardiac cell types in Drosophila at single cell resolution levels, we are investigating the signaling and transcriptional processes that determine their fates.
Examination of mesenchyme-to-epithelial transitions (METs).
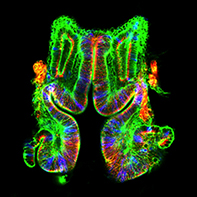
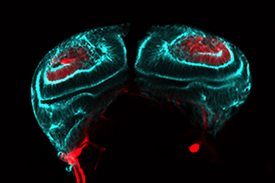 The transition from loosely associated, polarity-lacking mesenchymal cells to planar arrays of tightly-packed epithelial cells characterized by apical-basal polarity plays a critical role not only in mammalian cardiogenesis, but also in hepatogenesis (liver development) and nephrogenesis (kidney development). We are studying MET using a Drosophila imaginal disc model which exhibits a remarkable conservation of signals with mammalian kidney development.
The transition from loosely associated, polarity-lacking mesenchymal cells to planar arrays of tightly-packed epithelial cells characterized by apical-basal polarity plays a critical role not only in mammalian cardiogenesis, but also in hepatogenesis (liver development) and nephrogenesis (kidney development). We are studying MET using a Drosophila imaginal disc model which exhibits a remarkable conservation of signals with mammalian kidney development.
Investigating the transcriptional integration of multiple cues/inputs in gene regulation.
Many individual genes are regulated by multiple cues, such as sex, segmental identity, and tissue-specificity. We are exploring how these diverse cues are integrated at a transcriptional level.
